During my PhD, I took an active interest in science education, with some emphasis on undergraduate laboratory experience. I tried and incorporated projects which (1) would have piqued my interest as an undergraduate, (2) convey how important basic research is, in combination with medically relevant topics and (3) include a introductory bioinformatics experience.
Legacy Naturalist Badge Workshop
|
|
The Girl Scouts are committed to encouraging young ladies to pursue interests in the STEM fields. I was a Girl Scout for 8 years (Brownie to Cadette) and would love to give back to the Girl Scout community here in south Florida. Therefore, I have organized, in conjunction with the University of Miami and the Gifford Arboretum, a Naturalist Badge Day for Girl Scouts of several age groups.
Highlighted in the Spring 2015 issue of the University of Miami's College of Arts and Sciences Magazine, UM's eVeritas newletter, and by the Girl Scout Council of America. Volunteers: (Graduate Students) Kelley E, Andrea W, Winter B, Sarah C., Wyatt S. (Faculty 2014) Dr. Alex Wilson, Dr. Barbara Whitlock (UM) (Faculty 2015) Dr. James Baker, Dr. Carol Horvitz (UM) (Faculty 2016) Dr. Carol Horvitz (UM), Sara Zajic (Fairchild) |
Using a Single-Nucleotide Polymorphism to Predict Bitter-Tasting Ability

I introduced a half-semester long lab called "Using a Single-Nucleotide Polymorphism to Predict Bitter-Tasting Ability": curriculum included student discussions of primary scientific literature, performance of various laboratory techniques (DNA Extraction, PCR, Restriction Digest, Gel Electrophoresis), use of statistics in analysis of results (Chi-Squared), and an introduction to online bioinformatics tools (NCBI).
Class results will show how well PTC tasting actually conforms to classical Mendelian inheritance, and illustrate the modern concept of pharmacogenetics—where a SNP genotype is used to predict drug response.
This lab was introduced in Spring 2013, and is still a part of the Genetics Lab curriculum (as of Fall 2016).
Class results will show how well PTC tasting actually conforms to classical Mendelian inheritance, and illustrate the modern concept of pharmacogenetics—where a SNP genotype is used to predict drug response.
This lab was introduced in Spring 2013, and is still a part of the Genetics Lab curriculum (as of Fall 2016).
Studying the Evolution of the Domesticated Dog through Population Genetics and Bioinformatics
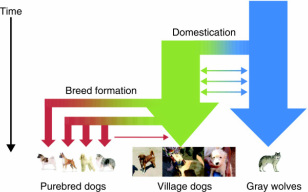
I put together a lab called "Studying the Evolution of the Domesticated Dog through Population Genetics and Bioinformatics": curriculum includes student led discussion of primary scientific literature, an active use of the biological statistics program Arlequin to analyze a data set and use of online bioinformatics tools to discover genes affected by domestication in dogs (NCBI).
This lab was crafted by me for the Honors Genetics Lab at the University of Miami. It includes a background information, plus a data set, provided by Dr. Adam Boyko, whose lab performed the actual research.
Part 1: Introduction to Genetic Markers, Part 2: Tracing the Domestication of Dogs using Molecular Markers, Part 3: Artificial Selection in Dogs (Village Dogs to Purebred Dogs)
Below is a zipped file with the (a) student copy (b) teacher copy (c) presentation PowerPoint and (d) data file. Please let me know if you have any questions.
This lab was crafted by me for the Honors Genetics Lab at the University of Miami. It includes a background information, plus a data set, provided by Dr. Adam Boyko, whose lab performed the actual research.
Part 1: Introduction to Genetic Markers, Part 2: Tracing the Domestication of Dogs using Molecular Markers, Part 3: Artificial Selection in Dogs (Village Dogs to Purebred Dogs)
Below is a zipped file with the (a) student copy (b) teacher copy (c) presentation PowerPoint and (d) data file. Please let me know if you have any questions.
|
If you have used this exercise, and have comments/suggestions, or even just want to tell me how it went, please email me!
graham.allie (a) gmail.com |
| ||||||
Silencing Genomes: Learning modern genetic techniques through RNAi
|
|
"Silencing genomes presents a series of experiments demonstrating the power of RNAi in the model organism, C.elegans. The curriculum begins with the observation of mutant phenotypes and basic worm "husbandry", progresses to simple methods to induce RNAi and to explore the mechanisms of RNAi using PCR, culminating with experimental methods to allow students to silence any gene in the C.elegans genome." http://www.silencinggenomes.org/
I introduced this series to the Miami-Dade College, as an authentic genetics research laboratory experience (1 semester - 15 weeks). It allowed students to utilize common laboratory techniques in the context of a hypothesis-driven project, and introduced them to bioinformatic tools commonly used in genetics research. Since this kit is no longer available, and I have made a fair number of modifications, please feel free to email me about details. |